- Richard West Awakening Through Change: A Practical Guide To Self-Discovery And Liberation In Times Of TransitionBinding : Taschenbuch, Label : Independently published, Publisher : Independently published, medium : Taschenbuch, numberOfPages : 171, publicationDate : 2018-12-02, authors : Richard West, ISBN : 1790645123
- Plug In Digital Of Orcs and MenLead the revolution of the Orcs and Goblins!In a world at war, the vast Empire of Men tries to extend its domination over the territories of the Orcs and Gob...
- Sarah Blaffer Hrdy Mothers And Others: The Evolutionary Origins Of Mutual UnderstandingBinding : Taschenbuch, Label : Harvard University Press, Publisher : Harvard University Press, NumberOfItems : 1, PackageQuantity : 1, medium : Taschenbuch, numberOfPages : 432, publicationDate : 2011-04-01, authors : Sarah Blaffer Hrdy, languages : english, ISBN : 0674060326
- Sycomore CRE6012 - Coloriage Enfant - Coloriages Villes 40 Pages - 3+ Ans SYCOMORE
- Coloriages Mystères Pixels Et Codes AnimauxBinding : Taschenbuch, Label : DEUX COQS D OR, Publisher : DEUX COQS D OR, Format : Illustriert, medium : Taschenbuch, numberOfPages : 96, publicationDate : 2024-02-14, ISBN : 2017247219
- Staedtler Feutres de coloriage double pointe Design Journey couleurs assorties - Pochette de 36 8Ces feutres de coloriage conviennent pour les croquis, les dessins et illustrations ainsi que l'écriture et le coloriage. Feutre de coloriage double pointe.<br/><br/>Corps en polypropylène.<br/>Largeur du tracé : 0,5 -3 mm.<br/><br/>Capuchon ventilé (ISO 11540). - Offre exclusivement réservée aux professionnels
- Staedtler Set 72 Feutres de coloriage double pointe fine 0,5 à 0,8mm et large 3mm, à base d'eau, assortisFeutres Design Journey<br/>Feutres double pointe.<br/>Pour de multiples applications : croquis, dessins, illustrations, écriture et coloriage, ...<br/>Encre à base d'eau et colorants alimentaires, lavable sur la plupart des textiles.<br/>Largeur de trait : pointe large 3 mm et pointe fine 0,5 à 0,8 mm.<br/>Conforme à la norme EN 71.<br/>Set en plastique rigide transparent pegboardable.<br/>Set de 72 feutres - Offre exclusivement réservée aux professionnels
- Ozzé Coloriages érotiques pour adultesJe suis le livre de Coloriages érotiques pour adultes de Ozzé. Original et divertissant, je suis un excellent cadeau à offrir à ses amis, par exemple pour un EVJF ou un EVJG ! Être adulte ne signifie pas ne plus être joueur ou joueuse...Je cache de nombreux coloriages au fil de mes pages, tous plus coquins les uns que les autres. Concentrez-vous pour colorier sans dépasser les lignes... Y arriverez-vous Dans tous les cas, vous passerez un bon moment !
- Staedtler Pochette de 10 feutres de coloriage double pointe de 2 couleurs et de 2 pointes différentes. - Lot de 3Feutre double pointe<br/>Etui de 10 feutres pegboardable en carton, composé à 80 % minimum de matériaux recyclés.<br/>Feutres de coloriage double pointe moyenne 1,0 mm et large 3,0 mm de 2 couleurs différentes soit 20 couleurs.<br/>Pointe bloquée solide et résistante à la pression.<br/>Encre à base d'eau et de colorants alimentaires, lavable sur la plupart des textiles.<br/>Produit éco-responsable :<br/>Recyclé à 80% - Offre exclusivement réservée aux professionnels
- Stabilo Feutre de coloriage pointMax, présentoir de 48Largeur de tracé: 0,8 mm, pointe nylon robuste, encre<br/><br/>inodore à base d'eau, protection contre le séchage pendant<br/><br/>24 heures, avec clip, capuchon enfichable au bout du feutre<br/><br/>contenu: 48 feutres<br/><br/><br/><br/>assorti dans les couleurs: 8x noir, 8x bleu outremer, 4x<br/><br/>vert émeraude, 4x carmin, 4x violet, 4x bleu turquoise,<br/><br/>4x vert feuille, 4x rouge rosé, 4x orange, 4x brun - Offre exclusivement réservée aux professionnels
- Stabilo Feutre de coloriage Trio Deco, étui en carton de 8 - Lot de 2Largeur du tracé: 2 - 3 mm, couleurs métalliques, zone de<br/><br/>grip triangulaire ergonomique, capuchon pouvant être fixé<br/><br/>sur l'embout du feutre<br/><br/>contenu: 8 pièces - Offre exclusivement réservée aux professionnels
- Bic Feutres de coloriage Visacolor XL ECOlutions® gigapack X288Kit Visacolor XL feutres + abécédaire<br/>Feutres de coloriage pointe Large.<br/>Maxi Schoolpack de 238 feutres + 50 gratuits + 1 Abécédaire à colorier pour apprendre l'alphabet.<br/>Couleurs assorties.<br/>Le kit - Offre exclusivement réservée aux professionnels
- Staedtler Feutre de coloriage triplus color,set anniversaire20 + 5, largeur du tracé: 1,0 mm, triangulaire, pointe résis<br/><br/>tante à la pression, encre à base d'eau, DRY SAFE, lavable,<br/><br/>à clip, capuchon ventilé, promo anniversaire 25 ans triplus,<br/><br/>contenu: 25 feutres (20 + 5 GRATUITS)<br/><br/><br/><br/>assorti dans les couleurs jaune, orange, rouge, bordeaux,<br/><br/>magenta, violet rouge, violet, bleu, bleu outremer, bleu<br/><br/>clair, bleu aqua, turquoise, vert, vert jaune, vert olive,<br/><br/>brun clair, brun, gris clair, gris, noir, jaune soleil,<br/><br/>rouge écarlate, saumon, vert pâle, bleu de Delft - Offre exclusivement réservée aux professionnels
- STABILO Pot de 48 feutres de coloriage pointe extra large Trio JumboSTABILO Trio Jumbo est un feutre de coloriage spécialement pensé pour les plus jeunes enfants à partir de 3 ans. Grâce à sa pointe large ultrarésistante à la pression et offrant un tracé de 3 mm, il va permettre aux plus petits de colorier rapidement de grandes surfaces. Sa zone de préhension triangulaire assure une prise en main confortable : dessiner, colorier et créer pendant des heures n'est plus un problème ! Et si dans un élan créatif l'enfant vient à se tacher, pas de panique : l'encre du feutre STABILO Trio Jumbo est super lavable sur la plupart des textiles, il suffit juste de mettre les vêtements à laver en machine avec votre lessive habituelle (température de lavage conseillée : 40°C). Regroupant 12 coloris assortis de STABILO Trio Jumbo, ce pot saura ranger et regrouper vos STABILO au même endroit.
- Pelikan Feutre de coloriage colorella, aquarellable, étui 6 - Lot de 3S'applique et se mélange au pinceau et à l'eau, lavable sur<br/><br/>la plupart des tissus, capuchon ventilé, pointe ogive,<br/><br/>largeur du tracé: env.<br/>2,0 à 43,0 mm<br/><br/>contenu : 6 feutres - Offre exclusivement réservée aux professionnels
- Staedtler Feutre de coloriage double pointe Noris,étui de 10 - Lot de 4Largeur de tracé: 1,0 et 3,0 mm, encre à base<br/><br/>d'eau et de colorants alimentaires, DRY-SAFE, pointes<br/><br/>blocables, lavable, capuchon ventilé, pointe ronde<br/><br/>contenu: 10 pièces<br/><br/><br/><br/>assortis dans les couleurs jaune, orange, rouge, violet,<br/><br/>bleu, bleu clair, vert, vert clair, marron, noir - Offre exclusivement réservée aux professionnels
- Staedtler Feutre de coloriage Noris, étui en carton 24 - Lot de 3Épaisseur mine: 1,0 mm, rond, facile à laver, capuchon avec<br/><br/>canal d'air, couleurs assorties<br/><br/>contenu: 24 pièces<br/><br/><br/><br/>assorti dans les couleurs jaune, jaune clair, jaune sable,<br/><br/>jaune doré, rouge, magenta, bordeau, bleu, bleu clair, bleu<br/><br/>de cobalt, turquois, orange, vert, vert-jaune, vert sève,<br/><br/>vert de terre, olive clair, vert olive, violet, violet<br/><br/>pourpre, marron, marron van-dyke, gris clair, noir - Offre exclusivement réservée aux professionnels
- Lot de 50 enveloppes carton B-Box 4 imprimée COLORIAGE format 250x353 mmPochette en carton plat blanc qualité 500 g/m² (crayons de couleurs non fournis) - Intérieur gris. - Excellente finition. - Rabat de fermeture autoadhésif avec bande de protection détachable. - Ouverture rapide et sécurisée ! - Idéal pour expédier livres, photos, agrandissements, diplômes, certificats, affiches, etc. - Permet d'insérer des objets d'une épaisseur jusqu'à 3,5 cm (objets dont les dimensions sont inférieures de 5 cm au format extérieur) - 100% recyclable. Le meilleur rapport qualité/prix en terme de protection et de résistance !
- BIC KIDS Kit de coloriage pour le voyage 'MEMORY GAME'Dans une mallette en métal, 64 pièces, dimensions de la<br/><br/>mallette: (L)207 x (P)157 x (H)71 mm, contenu: 12x crayon<br/><br/>de couleur triangulaire Evolutuion Triangle, 12x crayon cire<br/><br/>triangulaire Plasticolor Triangle, 8x feutre Visacolor XL,<br/><br/><br/><br/>jeu de mémory de 32 pièces à colorier - Offre exclusivement réservée aux professionnels
- Bic Ecriture Visaquarelle Feutres de Coloriage avec Pointe Pinceau - Couleurs Assorties, Pot de 48 OrangeFeutres Pinceaux Visaquarelle<br/>Feutres pointe extra souple indéformable et bloquée.<br/>Peut rester ouvert 4 semaines sans sécher ! Couleurs assorties.<br/>Pot de 48 - Offre exclusivement réservée aux professionnels
- STABILO Pot métal de 24 feutres de coloriage pointe large power maxRegroupant 12 coloris assortis de STABILO power max, ce pot saura ranger et regrouper vos STABILO au même endroit. Fabriqué en Europe, STABILO power max est le feutre de coloriage pointe large parfait pour les enfants dès 3 ans. Grâce à son corps large et rond, les plus petits à partir de 3 ans peuvent bien le prendre en main et son tracé de 3 mm permet à l'enfant de colorier rapidement de larges surfaces. Et comme il arrive d'oublier de remettre le capuchon, pas de problème il peut rester ouvert jusqu'à 8 semaines sans sécher! Sa pointe est bloquée pour résister à la pression et ainsi ne se casse pas et ne se tord pas. Son capuchon est ventilé, en cas d'ingestion et son encre est lavable sur la plupart des textiles.
- Staedtler Feutre de coloriage double pointe Noris,étui de 12 - Lot de 3Largeur de tracé: 1,0 et 3,0 mm, encre à base<br/><br/>d'eau et de colorants alimentaires, DRY-SAFE, pointes<br/><br/>blocables, lavable, capuchon ventilé, pointe ronde<br/><br/>contient: 12 feutres<br/><br/><br/><br/>assorti dans les couleurs: jaune, orange, rouge, violet,<br/><br/>carmin clair, bleu clair, bleu, vert, vert clair, marron,<br/><br/>noir, gris clair - Offre exclusivement réservée aux professionnels
- STABILO Schoolpack de 144 feutres de coloriage pointe moyenne Trio A-ZLe schoolpack est le format idéal pour les salles de classe : une grande quantité de feutres dans les 12 couleurs principales et une boîte de rangement super pratique. Les compartiments intégrés permettent de trier facilement les feutres par couleur, et une fois l'activité terminée on remet le couvercle et on range la boîte. Ce schoolpack est compatible avec certains meubles de rangement pour la classe. Le feutre STABILO Trio A-Z est le feutre idéal pour les petits artistes qui souhaitent un feutre de qualité pour écrire et dessiner. Ergonomique, sa zone grip triangulaire permet une prise en main détendue et un bon positionnement des doigts sur le feutre. Sa pointe moyenne de 0,7mm est parfaitement polyvalente : assez fine pour écrire, et assez large pour dessiner et colorier. Son encre est super lavable et permet aux taches accidentelles de partir après un lavage à 40°C sur la plupart des textiles. A l'école ou à la maison, le feutre STABILO Trio A-Z, disponible dans plus de 30 coloris dont 5 fluos, sera le compagnon créatif idéal des enfants.
- La Normandie Poster ColoriageBinding : Poster, Label : La Petite Boite, Publisher : La Petite Boite, PackageQuantity : 1, medium : Sonstige Einbände, publicationDate : 2015-07-21
- Dessin Et Coloriage Enfant Djeco Coffret Feutres Pinceaux 24 PiècesDessin et coloriage enfant Djeco Coffret feutres pinceaux 24 pièces - Dessin et coloriage enfant. Achat et vente de jouets, jeux de société, produits de puériculture. Découvrez les Univers Playmobil, Légo, FisherPrice, Vtech ainsi que les grandes marques de puériculture : , , Mac Laren, ...
- Staedtler Feutre de coloriage Noris, étui en carton 12 - Lot de 5Épaisseur mine: 1,0 mm, rond, facile à laver, capuchon avec<br/><br/>canal d'air, couleurs assorties<br/><br/>contenu: 12 pièces<br/><br/><br/><br/>assorti dans couleurs: jaune, rouge, magenta, bleu, bleu<br/><br/>clair, orange, vert, vert-jaune, violet, marron van-dyke,<br/><br/>gris clair, noir - Offre exclusivement réservée aux professionnels
- STABILO Schoolpack de 144 feutres de coloriage pointe moyenne powerLe schoolpack est le format idéal pour les salles de classe : une grande quantité de feutres dans les 12 couleurs principales et une boîte de rangement super pratique. Les compartiments intégrés permettent de trier facilement les feutres par couleur, et une fois l'activité terminée on remet le couvercle et on range la boîte. Ce schoolpack est compatible avec certains meubles de rangement pour la classe. Fabriqué en Europe, STABILO power est le feutre de coloriage pointe moyenne parfait pour les enfants qui oublient de remettre les capuchons ... Il peut rester ouvert jusqu'à 8 semaines sans sécher ! Sa pointe est bloquée pour résister à la pression et ainsi ne se casse pas et ne se tord pas. Disponible en 30 couleurs, son encre est lavable sur la plupart des textiles. Avec un tracé de 2 mm, STABILO power est le feutre pointe moyenne idéal pour la classe ou pour colorier et dessiner à la maison.
No statistical methods were used to predetermine sample size. The experiments that were randomized are indicated below and investigators were not blinded to allocation during experiments and outcome assessment, with the exception of the qPCR experiment, in which the technician at the IRIC-Genomics Platform was blinded to allocation during experiments and outcome assessment.
Ant culturing and collection
Colonies were maintained in plastic boxes with glass test tubes filled with water constrained by cotton wool, and were fed a combination of mealworms, crickets, fruit flies and Bhatkar–Whitcomb diet39. All colonies were maintained at 25 °C, 70% relative humidity and a 12-h day:night cycle.
Colonies were collected from the following locations: Aphaenogaster picea, Camponotus pennsylvanicus, Formica subscericea and Lasius niger were collected at McGill Gault Nature Reserve (Quebec, Canada) (45° 32′ 12.4′′ N, 73° 09′ 10.1′′ W ± 1 km). Camponotus novaeboracensis ants were collected at Winnipeg (Manitoba, Canada) (49° 51′ 12.6′′ N, 97° 08′ 14.0′′ W ± 1 km). Camponotus floridanus, Camponotus castaneus and Monomorium sp. were collected at Gainesville (Florida, USA) (29° 42′ 05.7′′ N, 82° 20′ 43.5′′ W ± 1 km). Colobopsis impressus ants were collected at Gainesville (Florida, USA) (29° 41′ 07.4′′ N, 82° 13′ 38.5′′ W ± 1 km). Camponotus ocreatus, Camponotus sansabeanus, Formica occulta and Veromessor pergandei were collected at Miami (Arizona, USA) (33° 24′ 28.1′′ N, 111° 00′ 14.5′′ W ± 1 km). Camponotus festinatus, Camponotus americanus, Camponotus sansabeanus, Brachymyrmex patagonicus, Nylanderia fulva and Nylanderia vividula were collected at University of Texas at Austin, Brackenridge Field Laboratory (Texas, USA) (30° 17′ 2.40′′ N, 97° 46′ 40.80′′ W ± 1 km). Myrmica americana and Prenolepis imparis were collected at Medford (New York, USA) (40° 48′ 6.8566′′ N, 73° 0′ 16.7756′′ W ± 1 km). Gigantiops destructor ants were collected at ACTS Research Station (Maynas, Peru) (3° 14′ 60.00′′ S, 72° 54′ 36.00′′ W ± 1 km). Anoplolepis gracillipes, Dolichoderus thoracicus, Oecophylla smaragdina, Paratrechina longicornis, Colobopsis leonardi and Polyrhachis rastellata were collected at Mae Tang (Chiang Mai, Thailand) (location data not available). Polyrhachis schlueteri were collected at Bela Bela (Limpopo, South Africa) (24° 47′ 32.0′′ S, 28° 17′ 30.6′′ E ± 1 km). Polyrhachis illaudata and Polyrhachis dives were collected at Hong Kong region Guangdong (China) (location data not available). Lasius emarginatus ants were collected at Palmanova (Udine, Italy) (45° 54′ 31.5′′ N, 13° 18′ 45.2′′ E ± 1 km).
Aphaenogaster picea, Camponotus pennsylvanicus, Formica subscericea, Lasius niger, Camponotus castaneus, Camponotus floridanus, Colobopsis impressus, Monomorium sp., Camponotus ocreatus, Camponotus sansabeanus, Formica occulta, Veromessor pergandei, Camponotus festinatus, Camponotus americanus, Camponotus sansabeanus, Brachymyrmex patagonicus, Nylanderia vividula, Myrmica americana and Prenolepis imparis were collected by the laboratory of E.A. Camponotus novaeboracensis was collected by J. Rand, Nylanderia fulva was collected by E. Lebrun and Gigantiops destructor was collected by J. Gibson (laboratory of A. Suarez). Anoplolepis gracillipes, Dolichoderus thoracicus, Oecophylla smaragdina, Paratrechina longicornis, Colobopsis leonardi and Polyrhachis rastellata were purchased from Ants of Asia (P. Williams), and Polyrhachis schlueteri, Polyrhachis illaudata, Polyrhachis dives and Lasius emarginatus were purchased from Antstore (M. Sebesta).
Ovary dissections
This protocol was modified from a previous publication40. Ovaries were dissected in 0.1% PBSTween (1.86 mM NaH2PO4, 8.41 mM Na2HPO4, 1.75 M NaCl, 0.1% Tween20, pH 7.4) and kept on ice until fixation. First, the ovaries were removed from the oviduct. Ovaries were then separated into individual ovarioles, and the peritoneal sheath was then removed with fine forceps. Ovarioles were fixed in a solution of 5% formaldehyde (135 μl), 10% DMSO (100 μl) in 0.1% PBSTween (765 μl) for 25 min at room temperature, then washed with 0.1% PBSTween and gradually transferred to a solution of 100% methanol for storage.
Embryo collection and fixation
This protocol was modified from previous publications40,41,42. Embryos were treated with 4% hypochlorite solution (bleach) for 2 min. Embryos used for immunohistochemistry were then fixed using a ‘slow formaldehyde fixing method’ using PEMS (100 mM PIPES, 2 mM MgSO4, 1 mM EGTA, pH 6.9) and were treated with proteinase K (New England Biolabs) in PBS at a final concentration of 0.08 U/ml. Embryos used for in situ hybridization were heat-fixed using a boiling hot solution of PBS-Triton (1.86 mM NaH2PO4, 8.41 mM Na2HPO4, 1.75 M NaCl, 0.03% Triton-X-100, pH 7.4).
Embryo staging
Timed egg depositions were collected in smaller setups and allowed to develop at 25 °C, 70% relative humidity along with a few workers, and fixed at two-hour windows. The embryos were DAPI-stained and observed under differential interference contrast (DIC) and wide-field fluorescence for staging. As far as possible, the staging scheme landmarks used correspond to Bownes’ staging scheme for Drosophila43.
Whole-genome shotgun sequencing
Whole DNA was isolated from 0–6-h-old embryos using Qiagen Genomic-tip 20/G kit. Shotgun sequencing was performed at Genome Quebec using Illumina HiSeq platform. Sequences were curated and BLAST searches performed using Geneious software44.
Gene cloning and molecular biology
Gene sequences were obtained from NCBI GenBank database using genome BLAST against the assembled C. floridanus genome45. The accession numbers of genes used in this study are: abdA XM_020027891.2; nos XM_011266396; osk XM_011254572.2; smg XM_011254071.3; stau XM_011254361.3; Ubx XM_011259757.1; en XM_011252307.3. It was necessary to use a better-annotated Ubx cDNA sequence, which was submitted to GenBank under accession number MH801205. Camponotus floridanus and L. niger RNA was isolated using TRIzol (Invitrogen) from a pool of embryos and larvae of different developmental stages. RNA was then reverse-transcribed to synthesize a cDNA library. Specific primers were designed to amplify the gene fragments from cDNA libraries prepared from embryos and cloned in pGemT-easy vector (Promega) using standard procedures, and subsequently sequenced using Sanger sequencing at the Genome Quebec Innovation Centre. The primers used were: osk forward 5′-CGGAGAGCCTATTCCTTATC- 3′, and reverse 5′-GCCAGAGATCTGATCCAATTA- 3′, nos forward 5′-TCCCAGTTTGGACGAAGAATAAAG- 3′, and reverse 5′-GTTTTCCCGCAGAGTTTCTCAGTA- 3′, stau forward 5′-GCGAATTCACGGGTAGAGGT- 3′, and reverse 5′-GAAACACCAGCCGCATTCTG- 3′, abdA forward 5′-GTCTTCCTAAGAGCGACGAGC- 3′, and reverse 5′-GTGGGTACCTTACTGACTGCC- 3′, Ubx forward 5′-GCTTCTACGGAAGCCACCATC- 3′, and reverse 5′-TGCTTCTCCTGCTCGTTTAGC- 3′, smg forward 5′-TCACTTTTGCGTCGTCTACCT- 3′, and reverse 5′-AGAGAGAGCCAGTTTGTGCC- 3′, en forward 5′-CGACACGAGCGAGGTATTGA- 3′, and reverse 5′-GAGGCCGATCGATTTGACGA- 3′.
Identifying orthologues and paralogues
Amino acid sequence alignments were done using ClustalW in Geneious platform confirming the orthology of vasa (vas), oskar (osk), nanos (nos), tudor (tud), germ cell-less (gcl), staufen (stau), caudal (cad), smaug (smg), wunen-2 (wun2), aubergine (aub), heat shock protein 90 (hsp90), argonaute 3 (ago3), abdominal A (abdA), Ultrabithorax (Ubx) and engrailed (en). The alignments are presented in Supplementary Figures 1–15. To search for any lineage-specific paralogues of the germline genes that we studied, a blastn search was performed on the latest C. floridanus genome assembly accessed from: https://www.ncbi.nlm.nih.gov/assembly/1752781 using a maximum E value of 0.05; scoring of 2, −3; and gap cost of 5, 2. Only hits above an e-value cut off (e−20) were considered and highlighted in bold. If the query subject had more than one hit but aligned to the same contig number, then it was concluded that no paralogues for the gene in question exist. However, if the query subject had more than one hit but was aligned to multiple contig numbers, then it was concluded that paralogues for the gene in question do exist. The hit tables are presented as Supplementary Table 3.
Immunohistochemistry and in situ hybridization
The following are primary antibodies we used in this study, the concentrations at which we used them at, and their source: mouse anti-HSP90 (1:100) antibody (BD bioscience 610418) and mouse anti-UbdA (1:4) antibody (FP6.87, DSHB), rabbit anti-Vasa (1:100) antibody (gift from P. Lasko), rabbit anti-Tudor (1:100) antibody (gift from P. Lasko), rabbit anti-Germ cell-less (1:300) antibody (gift from P. Lasko), rabbit anti-Aubergine (1:50) antibody (gift from P. Lasko), and rabbit anti-Oskar (1:100) antibody (gift from P. Lasko). Fluorescent secondary donkey anti-rabbit and anti-mouse polyclonal Alexa Fluor-488 (AbCam) antibodies were used at 1:500 dilution to detect the primary antibody, according a previous publication30. In situ hybridization was done according to previous publications40,46, modified for in situ robot InsituPro VSi (Intavis) with the following modifications; the duration of wash steps was maintained according to the cited protocol but the buffer was exchanged every 5 min to increase agitation. Alkaline phosphatase secondary antibody anti-DIG-AP (Roche) was used to detect DIG-labelled probes and streptavidin-AP (Roche) reagent was used to detect biotin-labelled probes. Templates for probes were prepared using PCR with T7 and SP6 primers on the plasmids containing cloned gene fragments. Probe synthesis was done using SP6 or T7 RNA polymerase (Roche) according to the suppliers’ directions. Probes were purified using phenol–chloroform and isopropanol precipitation method according to a previous publication47, and used at 3 ng/μl final concentration. The probes consisted of 538 bp of abdA (bases 28–566), 424 bp of nos (bases 51–475), 848 bp of osk (bases 96–944), 987 bp of stau (bases 1121–2108) and 874 bp of Ubx (bases 32–906), 1,037 bp of en (bases 201–1237) and 949 bp of smg (bases 268–1197), in which base numbering starts at the start codon.
Microinjections and RNAi for phenotypic analysis
Embryos were collected as timed depositions from queens isolated with at least a dozen minor workers and at least six larvae and pupae. To eliminate any colony or day-of-injection related effects, embryos from multiple queens were collected, randomized between treatment and control and injected on the same day. Embryos were lined up alongside a fine glass capillary on a Petri dish lid lined with a thin layer of 2% agar in water and supplemented with 10 μl of 10 μg/ml ampicillin, modified from a previous publication48. Injection needles were prepared using a micropipette capillary puller. Microinjections were done using FemtoJet Express and InjectmanNI2 (Eppendorf) setup on a Zeiss Axiovert zoom inverted microscope using the following settings: control pressure 2 psi, injection pressure 18 ps, and injection time 0.1 s. The needle tip was broken open by gently pushing it against a glass coverslip immersed in halocarbon oil. The injection volume was adjusted after the needle was broken. Injection volumes were between 0.5 and 1 nl. Embryos were incubated at 25 °C 70% relative humidity chamber. Embryos were transferred every 24 to 48 h on to fresh 50-mm Petri-dishes containing 2% agar in water topped with a Whatmann filter paper and supplemented with 10 μl of 10 mg/ml ampicillin. DNA templates for double-stranded (ds) RNA were prepared using PCR with M13 forward universal primer and M13 reverse universal primer containing a T7 promoter overhang on plasmids containing cloned gene fragments as templates. The templates were used to generate dsRNA using T7 RNA polymerase (Roche) according to manufacturer’s instructions. For controls, dsRNA was generated using the same method from a plasmid containing cloned 720 bp of the YFP coding sequence.
Quantitative PCR
Microinjections and RNAi
Embryos were collected as timed depositions from queens isolated with at least a dozen minor workers and at least six larvae and pupae. To eliminate any colony or day-of-injection related effects, embryos from multiple queens were collected, randomized between treatment and control and injected on the same day. Embryos were lined up alongside a fine glass capillary on a Petri dish lid lined with a thin layer of 2% agar in water and supplemented with 10 μl of 10 μg/ml ampicillin, modified from a previous publication48. Injection needles were prepared using a micropipette capillary puller. Microinjections were done using FemtoJet Express and InjectmanNI2 (Eppendorf) setup on a Zeiss Axiovert zoom inverted microscope using the following settings: control pressure 2 psi, injection pressure 18 psi and injection time 0.1 s. The needle tip was broken open by gently pushing it against a glass coverslip immersed in halocarbon oil. The injection volume was adjusted after the needle was broken. Injection volumes were between 0.5 and 1 nl. Embryos were incubated at 25 °C 70% relative humidity chamber. Embryos were transferred every 24 h on to fresh 50-mm Petri dishes containing 2% agar in water topped with a Whatmann filter paper and supplemented with 10 μl of 10 mg/ml ampicillin. DNA templates for dsRNA were prepared using PCR with M13 forward universal primer and M13 reverse universal primer containing a T7 promoter overhang on plasmids containing cloned gene fragments as templates. The templates were used to generate dsRNA using T7 RNA polymerase (Roche) according to manufacturer’s instructions. For controls, dsRNA was generated using the same method from a plasmid containing cloned 720 bp of the YFP coding sequence.
Sample preparation for qPCR
Embryos were collected at stage 8, 5 days after injection and heat-fixed by immersing in a boiling hot solution of PBS-Triton (1.86 mM NaH2PO4, 8.41 mM Na2HPO4, 1.75 M NaCl, 0.03% Triton-X-100, pH 7.4) for 1 min followed by rinses with ice-cold PBS. Individual germline capsules, bacteriocytes and yolk sacs with intact germbands curled around them were separated using sharpened tungsten needles, and extraembryonic serosa tissue was discarded. For each gene, 40 individual samples were divided into 4 technical replicates of 10, and the 10 samples within each technical replicate were pooled and immediately placed in 200 μl Trizol reagent. RNA was prepared on the same day using standard Trizol method. First-strand cDNA synthesis was done using Superscript-II reverse transcriptase (ThermoFisher). Instead of universal oligo-dT primers an equimolar pool of the following 21 gene specific primers (including those of 8 endogenous control genes) was used to account for low yields in small tissue preparations: vasa 5′-CGATATCTGGTAGAAAGCCC- 3′, osk 5′-GCCAGAGATCTGATCCAATTA- 3′, nos 5′-GTTTTCCCGCAGAGTTTCTCAGTA- 3′, tud 5′-AGCGCCGGTTCTATCATGTC- 3′, gcl 5′-CCATCTCCAAGTATGTTCACC- 3′, stau 5′-GAAACACCAGCCGCATTCTG- 3′, smg 5′-AGAGAGAGCCAGTTTGTGCC- 3′, ago3 5′-TACACCCGTTATGCTTTTGA- 3′, cad 5′-AGAGGCGCCGATAGAGATGAA- 3′, arm 5′-TCTCGGTGCCTGTGATTCTG- 3′, abdA 5′-TCCAGGCCGCTTACGTGATG- 3′, Ubx 5′-TGCTTCTCCTGCTCGTTTAGC- 3′, wun2 5′-TCGTAATCGGTAGGTCGATGC- 3′, act5c 5′-GAACGGTGTTGGCGTACAGA- 3′, tub 5′-CGACGGAGAGTTGTTCGTGA- 3′, argk 5′-CCTGTCCAAGATCACCACCC- 3′, ef1 5′-AGTGGTCAATCCAGCAGGTG- 3′, efl-like 5′-GCAGCTGGTATTCCCGTTTG- 3′, hisH-3 5′-CCCTGAAAAGGGCCGATTGT- 3′, rp60S 5′-AACGTGCACTGGCATTTGTC- 3′, and gadph 5′-ATTCGCCATACGACGAGACC- 3′.
Quantitative PCR
Quantitative PCR was performed at IRIC-Genomics Platform using qPCR Taqman method49 with the following primers: vasa forward 5′-CACAAGTACTTATTGTATCACCCACA-3′and reverse 5′-GAAAATTTCTTGGCCTGTTGA-3′, osk forward 5′-AATCTCGTCGGAGAGCCTAT-3′and reverse 5′-AAATGCACGGAGACTCGAAA-3′, nos forward 5′-CCTTACCAACAGAATGCGTCT-3′and reverse 5′-TCCTTTAGCAGATGTTTTCGATAG-3′, tud forward 5′-ATTGTGGGTACGAATATGTTATCG-3′and reverse 5′-ATGACAATGGTGTTAACATAAAGGAT-3′, gcl forward 5′-AAAACGATGGTTGGAAGTCAA-3′and reverse 5′-TGCCATTAAATCTGGTGCAA-3′, stau forward 5′-AACCCGCGAAACCATCTAT-3′and reverse 5′-CGTCACTTTTCTGGGTTTCG-3′, ago-3, forward 5′-TGGCATAGATGTCTATCATGCTG-3′and reverse 5′-GCAACAAATCCTGCAACACTC-3′, cad forward 5′-ATGTCAATGCAGGCAGCAC-3′and reverse 5′-ACGTGGACGGAGATGTCG-3′, wun2 forward 5′-TCTTGGCACAATCGTAGCTTT-3′and reverse 5′-TCCGTGGAAGAATGCCTCT-3′, tub forward 5′-CACAGGCACGTATCGACAAC-3′and reverse 5′-GCCACGCGCATAATTGTT-3′, act5c forward 5′-CGTCATCAGGGTGTCATGG-3′and reverse 5′-CAAGATACCTCTCTTCGATTGAGC-3′, rp60S forward 5′-GCGTTTCAAGGGCCAATAC-3′and reverse 5′-GCAGCATGTGACGTGTTTTC-3′, argk forward 5′-TGGTAGACGCAGCGGTTT-3′and reverse 5′-AACGACTTGCTGTCGGATTC-3′, efl-like forward 5′-ACGTTATTGTCGAGGCCAAG-3′and reverse 5′-GGCAGGACGTATCTGCGTA-3′, ef1 forward 5′-GCTGCAGTCGCATTTGTTC-3′and reverse 5′-ATCTTGGAAGATGGCTCCAG-3′, gapdh forward 5′-GCGGTGCCAAGAAGGTTAT-3′and reverse 5′-CCAAGTTTACACCGACAACG-3′, hisH-3, forward 5′-CTACTAAAGCGGCGAGGAAG-3′and reverse 5′-CCAGGCCTATAACGATGAGG-3′.
The endogenous control genes used (the last 8 primer pairs above) were: Actin5c, 60S ribosomal protein, arginine kinase, efl-like, elongation factor1, gadph, histone H3 and tubulin. The most stable endogenous controls were established through the use of the algorithms integrated in the RefFinder package50 that integrates four different protocols; GeNorm, BestKeeper, NormFinder and the comparative ΔCT method51,52,53,54 (Supplementary Fig. 16). Four of the endogenous controls (gapdh, hisH3, rp60S and argk) were deemed most stable and the geometric mean of these was used for calculating ΔCT values for each target gene within each biological sample and replicate according to previously published recommendations51 (Supplementary Fig. 16). Relative quantifications for abdA RNAi, Ubx RNAi and control YFP RNAi were calculated by the formula: relative quantification = 2−ΔΔCT, in which ΔΔCT is the difference between ΔCT in each RNAi sample and the average of ΔCT values in all YFP RNAi replicates of that treatment group. ΔΔCT values for each of the individual data points of the control YFP RNAi were also calculated using the average of all YFP RNAi from that particular biological replicate (black bars in Extended Data Fig. 4q, r). This method allows for consistency because the statistical analyses are performed on the same relative quantification values that are used to plot the bar graphs.
Antibiotic treatment
Two mature colonies were treated with rifampicin to test the effects of Blochmannia on embryonic development of C. floridanus. Rifampicin powder (Sigma; R883) was dissolved in water at a stock concentration of 2 mg/ml and then diluted 1:1 (final concentration 1 mg/ml rifampicin) in a 50% honey–water (Kirkland Signature) solution. Colonies were given fresh rifampicin–honey–water three times a week for two months. After two months, embryos were collected, fixed and stained with DAPI to confirm elimination of Blochmannia. Once elimination of Blochmannia was confirmed, embryos were collected and fixed for subsequent gene expression analysis. To rule out the possibility that the changes in phenotypes and gene expression or localization observed after rifampicin treatment are the unspecific effect of antibiotics were performed two controls: (1) a C. floridanus colony was treated with ampicillin, which does not eliminate Blochmannia from the colonies. Ampicillin powder (Fisher scientific; BP1760-25) was dissolved in water at a stock concentration of 400 mg/ml and then diluted 1:1 (final concentration 200 mg/ml ampicillin) in 50% honey–water solution. Colonies were treated in exactly the same manner as that for rifampicin. (2) An L. niger colony—a species that is in the same subfamily as C. floridanus but lacks Blochmannia—was treated with rifampicin in the same manner as C. floridanus. Lasius niger colonies were also treated with the same rifampicin regimen as C. floridanus and embryos were collected and fixed for subsequent gene expression analysis after at least two months.
Phylogenetic sampling, developmental characters and ancestral state reconstruction
Phylogenetic sampling
Thirty-one ant species were sampled in total: 26 from the subfamily Formicinae and 5 from 2 sister subfamilies of the Formicinae, the Myrmicinae (4 species) and the Dolichoderinae (1 species). Within the Formicinae, 14 in-group species within the Camponotini that evolved the obligate endosymbiosis with Blochmannia were sampled, and 12 out-group species were sampled that lack Blochmannia. Phylogenetic relationships and branch length information for these 31 species were obtained from previous molecular phylogenetic studies26,55,56,57.
Developmental characters
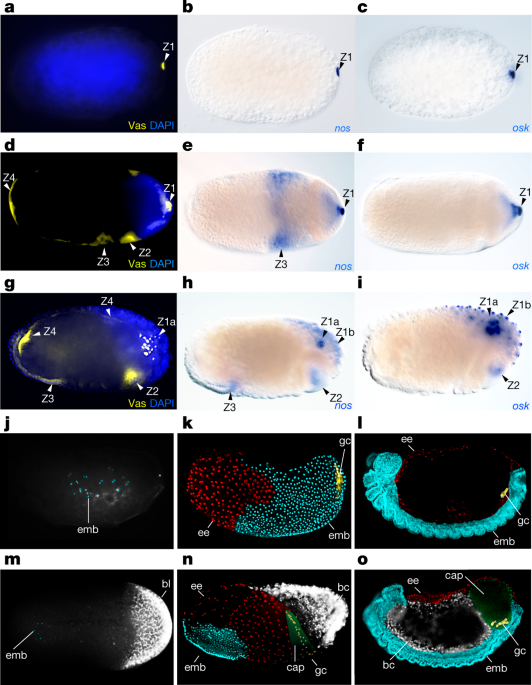
The following five developmental characters were characterized for each species: (1) character 1 is defined as the presence of specific localization zones of mRNAs and proteins of the germline genes based on Vas protein. Character 1 has four states: an embryo with the presence of 1, 2, 3 or 4 localization zones of mRNAs and proteins of germline genes as illustrated in Fig. 4a; (2) character 2 is defined as the presence of specific localization zones of mRNAs and proteins of the maternal Hox genes Ubx and abdA based on the UbdA antibody that recognizes both Ubx and AbdA protein (with the exception of 1 species, C. impressus, which is based on abdA mRNA). Character 2 has four states: an embryo with the presence of 0, 1, 3 or 4 localization zones of mRNAs and proteins of maternal Hox genes Ubx and abdA, as illustrated in Fig. 4a; (3) character 3 is defined as the presence and type of obligate endosymbionts at the posterior of the egg on the basis of our own data and previous studies8,9,58,59. Previous phylogenetic evidence10,25,38 showed that the three types of obligate endosymbionts within the Formicinae—the Camponotini obligate endosymbiont (Blochmannia), the Formica obligate endosymbiont and the Plagiolepidini obligate endosymbiont—were acquired independently and evolved convergently. Therefore, character 3 has four different states: an obligate endosymbiont at the posterior is absent; the Camponotini obligate endosymbiont (Blochmannia) is present at the posterior of the egg; the Formicini obligate endosymbiont is present at the posterior of the egg; or the Plagiolepidini obligate endosymbiont is present at the posterior of the egg, as illustrated in Fig. 4a; (4) character 4 is defined as the location of the embryo within the egg. Character 4 has two states: either the embryo is located in the posterior of the egg or the embryo is located in the anterior of the egg, as illustrated in Fig. 4a; (5) character 5 is defined as the germline capsule. Character 5 has two states: either the germline capsule is present or the germline capsule is absent, as illustrated in Fig. 4a.
Ancestral state reconstruction
RevBayes (v.1.7.10)37 was used to reconstruct ancestral character states for the 5 developmental characters across 31 ant species sampled. RevBayes37 uses Bayesian Markov chain Monte Carlo (MCMC) methods to estimate model parameters60. Ancestral states were estimated using two evolutionary models for discrete characters: the ‘equal-transition rates’ and ‘unequal-transition rates’ models. The equal-transition rates model assumes characters are equally likely to change from any one state to any other state, whereas the unequal-transition rates model assumes that the transition from any one state to any other state is unequal and can occur according to different rate parameters37. Both models were applied on each of the five developmental characters, and each model was run independently twice for 1,000,000 MCMC generations sampling every 500 generations. After completion of the MCMC analysis, the first 25% of the trees were discarded as a burn-in. Convergence between chains, likelihood scores and estimate sample size values were evaluated using Tracer (version 1.7)61. The estimate sample size value for each parameter sampled from the MCMC analysis was always recorded as >1,000, indicating that the number of effectively independent draws from the posterior distribution from of all MCMC runs was adequate. Model selection was performed using marginal log-likelihoods, which represent the probability of the data given a specific model integrated over all possible parameter values37. Bayes factors were computed and used to estimate and compare the probabilities of the unequal and equal models given the data for each developmental character. Stepping-stone sampling (50 MCMC runs in RevBayes37) was used to approximate the marginal log-likelihoods62. The unequal model was found to be the model that best fit the data for all developmental characters (Extended Data Table 2). Nonetheless, the equal model also gives posterior probabilities similar to those of the unequal model (Extended Data Table 2), indicating that the reconstruction obtained for each dataset is robust to the evolutionary model assumed. Finally, we assessed the sensitivity of these posterior probabilities to the branch lengths obtained from the literature by repeating all of the above analyses, but setting all branch lengths equal to 1. The posterior probabilities obtained with all branch lengths equal to 1 were similar to those obtained from the literature (Supplementary Table 2), indicating that the reconstruction obtained for each dataset is robust to the branch lengths used.
Microscopy
We used a Zeiss Discovery V12 stereomicroscope and Zeiss Axiovision software to image embryos and ovaries. For high-resolution imaging, we used Leica SP8 confocal microscope. ImageJ2 was used for analysis of images63.
Statistics and reproducibility
For a given gene, in situ hybridization and immunohistochemistry, the sample size for C. floridanus consisted of at least 30 embryos or ovarioles of similar stages; for other species that produce far fewer embryos, the sample size consisted of at least 5 embryos of similar stages. One hundred per cent of the embryos sampled showed the same expression patterns. In situ hybridization and immunohistochemistry experiments for C. floridanus were repeated at least eight times independently. For other species, these experiments were repeated at least four times. For RNAi experiments, phenotypes were considered reproducible if at least three independent replicates gave the same results. For qPCR, statistical analysis was performed using Graphpad Prism v7. or Microsoft Excel. Relative quantification values for YFP RNAi,s abdA RNAi and Ubx RNAi were calculated by the same method to ensure consistency between plotted results on the graph and for analysis of variance (ANOVA). Two-way ANOVA with replication was performed, in which Ubx RNAi was compared with YFP RNAi and abdA RNAi with YFP RNAi, treating RNAi as fixed and nine target genes as random effects. Each of the tissues (zone 1, zone2 and zone 3 + zone 4) was analysed by ANOVA as a separate experiment. The qPCR experiments were performed blind at the Genomic Platform facility at the Institute for Research in Immunology and Cancer. Fisher’s exact test was performed to determine: (i) whether there is a significant difference in phenotype frequency (wild-type-like versus mild or severe) between control embryos collected from rifampicin-treated colonies versus tranplanted embryos collected from rifampicin-treated colonies. Analyses were considered statistically significant at α < 0.05. For blinding and reproducibility, two different researchers independently performed the following steps without communicating each step: sample collection from colonies, randomization of embryos between treatments, treatment of samples, replicate maintenance, and data acquisition and analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.